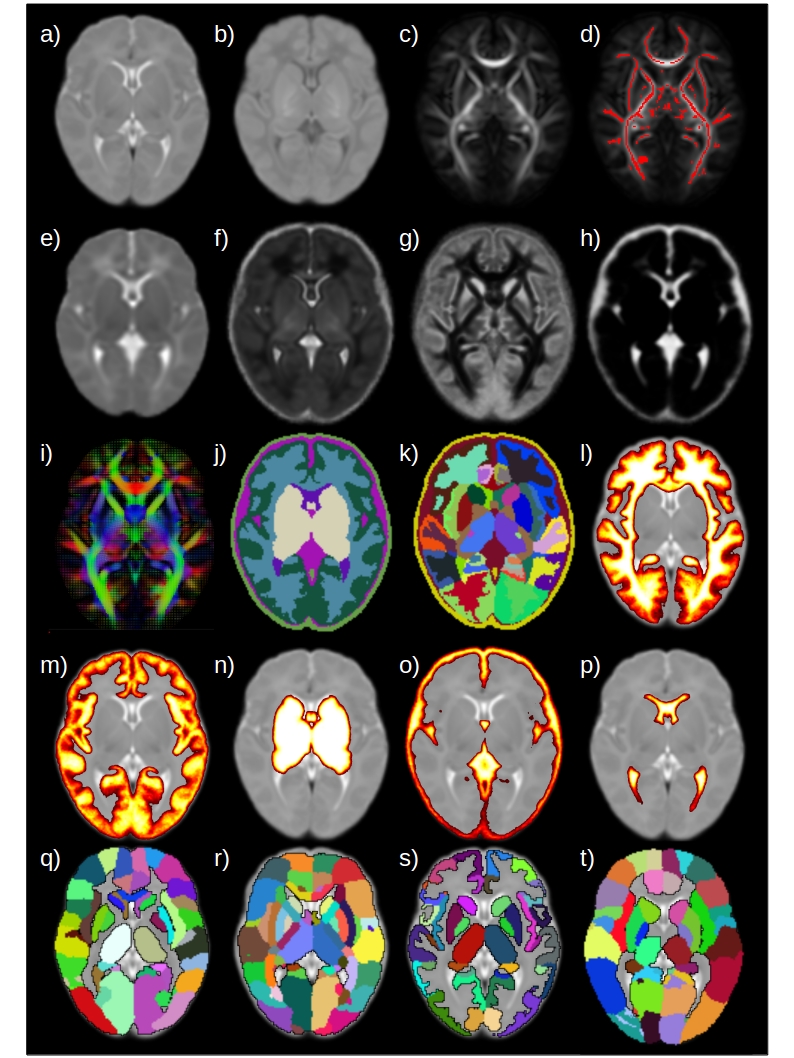
Edinburgh Neonatal Atlas
In this repository you can find all the versions of the Edinburgh Neonatal Atlas (ENA). The available versions are:
- ENA33: Blesa, M., Serag, A., Wilkinson, A. G., Anblagan, D., Telford, E. J., Pataky, R., Sparrow S.A., Macnaught, G., Semple S. I., Bastin M. E. and Boardman J. P. (2016). Parcellation of the healthy neonatal brain into 107 regions using atlas propagation through intermediate time points in childhood. Frontiers in Neuroscience 10, 220. doi:https://doi.org/10.3389/fnins.2016.00220
- ENA50: Blesa, M., Galdi, P., Sullivan, G., Wheater, E. N., Stoye, D. Q., Lamb, G. J., Quigley, A. J., Thrippleton, M. J., Bastin, M. E. and Boardman, J. P. (2020). Peak width of skeletonized water diffusion MRI in the neonatal brain. Frontiers in Neurology, 11, 235. doi:https://doi.org/10.3389/fneur.2020.00235
We encourage the user to try what atlas better fits their needs, but the general advice is to use the ENA50. The ENA50 is an improved version of the ENA33 with the following properties:
- Includes more templates (T1w, T2w, tensor template, DTI metrics templates and NODDI metrics templates).
- Different parcellation schemes (ENA33, JHU, UNC, M-CRIB and dHCP).
- More tissue probability maps.
- Improved alignemt between modalities (Due to the improved skull-stripping algorithm derivated from the dHCP framework and the use of TOPUP and EDDY).
All the parcellations were propagated to the ENA50 space for convenience (specially thinking in future studies that requiere normalization to a common space), but if you plan to use the additional parcellation schemes for other porpuses (such as connectivity or volumetric studies) is better to use the original templates (with the exception of the ENA33, where the ENA50 should be preferably used).
White Matter tracts
A set of ROIs were defined to create a set of WM tracts. The tracts of interest are:
- AF left/right
- ATR left/right
- CC genu
- CC splenium
- CCG left/right
- CST left/right
- IFOF left/right
- ILF left/right
- UNC left right
For more details about how the tracts are generated and how to use this set of ROIs, look at the following publication:
- Vaher, K., Galdi, P., Blesa, M., Sullivan, G., Stoye, D. Q., Quigley, A. J., Thrippleton, M. J., Bogaert, D., Bastin, M. E., Cox, S. R. and Boardman, J. P. (2021). General factors of white matter microstructure from DTI and NODDI in the developing brain. NeuroImage. doi: https://doi.org/10.1016/j.neuroimage.2022.119169
The scripts to generate the tracts can be found here: https://git.ecdf.ed.ac.uk/jbrl/neonatal-gfactors
Referencing
Please, if you use any of this templates, please cite all the corresponding work:
- ENA33 templates and parcellation:
- Blesa, M., Serag, A., Wilkinson, A. G., Anblagan, D., Telford, E. J., Pataky, R., Sparrow S.A., Macnaught, G., Semple S. I., Bastin M. E. and Boardman J. P. (2016). Parcellation of the healthy neonatal brain into 107 regions using atlas propagation through intermediate time points in childhood. Frontiers in Neuroscience 10, 220. doi:https://doi.org/0.3389/fnins.2016.00220
- ENA50 templates:
- ENA50: Blesa, M., Galdi, P., Sullivan, G., Wheater, E. N., Stoye, D. Q., Lamb, G. J., Quigley, A. J., Thrippleton, M. J., Bastin, M. E. and Boardman, J. P. (2020). Peak width of skeletonized water diffusion MRI in the neonatal brain. Frontiers in Neurology, 11, 235. doi:https://doi.org/10.3389/fneur.2020.00235
- Parcellation schemes:
- JHU: Oishi, K., Mori, S., Donohue, P. K., Ernst, T., Anderson, L., Buchthal, S., et al. (2011). Multi-contrast human neonatal brain atlas: Application to normal neonate development analysis. NeuroImage 56, 8 – 330 20. doi:https://doi.org/10.1016/j.neuroimage.2011.01.051
- UNC: Shi, F., Yap, P.-T., Wu, G., Jia, H., Gilmore, J. H., Lin, W., et al. (2011). Infant brain atlases from neonates to 1- and 2-year-olds. PLOS ONE 1–11. doi:https://doi.org/10.1371/journal.pone.0018746
- M-CRIB: Alexander, B., Murray, A. L., Loh, W. Y., Matthews, L. G., Adamson, C., Beare, R., et al. (2017). A new neonatal cortical and subcortical brain atlas: the melbourne children’s regional infant brain (m-crib) atlas. NeuroImage 147, 841 – 851. doi:https://doi.org/10.1016/j.neuroimage.2016.09.068
- dHCP: Makropoulos, A., Robinson, E. C., Schuh, A., Wright, R., Fitzgibbon, S., et al. (2018). The developing human connectome project: A minimal processing pipeline for neonatal cortical surface reconstruction. NeuroImage 173, 88–112. doi:https://doi.org/10.1016/j.neuroimage.2018.01.054 AND Makropoulos, A., Aljabar, P., Wright, R., Huning, B., Merchant, N., et al. (2016). Regional growth and atlasing of the developing human brain. NeuroImage 125, 456–478. doi:https://doi.org/10.1016/j.neuroimage.2015.10.047
- WM tracts:
- Vaher, K., Galdi, P., Blesa, M., Sullivan, G., Stoye, D. Q., Quigley, A. J., Thrippleton, M. J., Bogaert, D., Bastin, M. E., Cox, S. R. and Boardman, J. P. (2021). General factors of white matter microstructure from DTI and NODDI in the developing brain. NeuroImage. doi: https://doi.org/10.1016/j.neuroimage.2022.119169
Contact
For any doubt/suggestion please email me to: manuel.blesa@ed.ac.uk